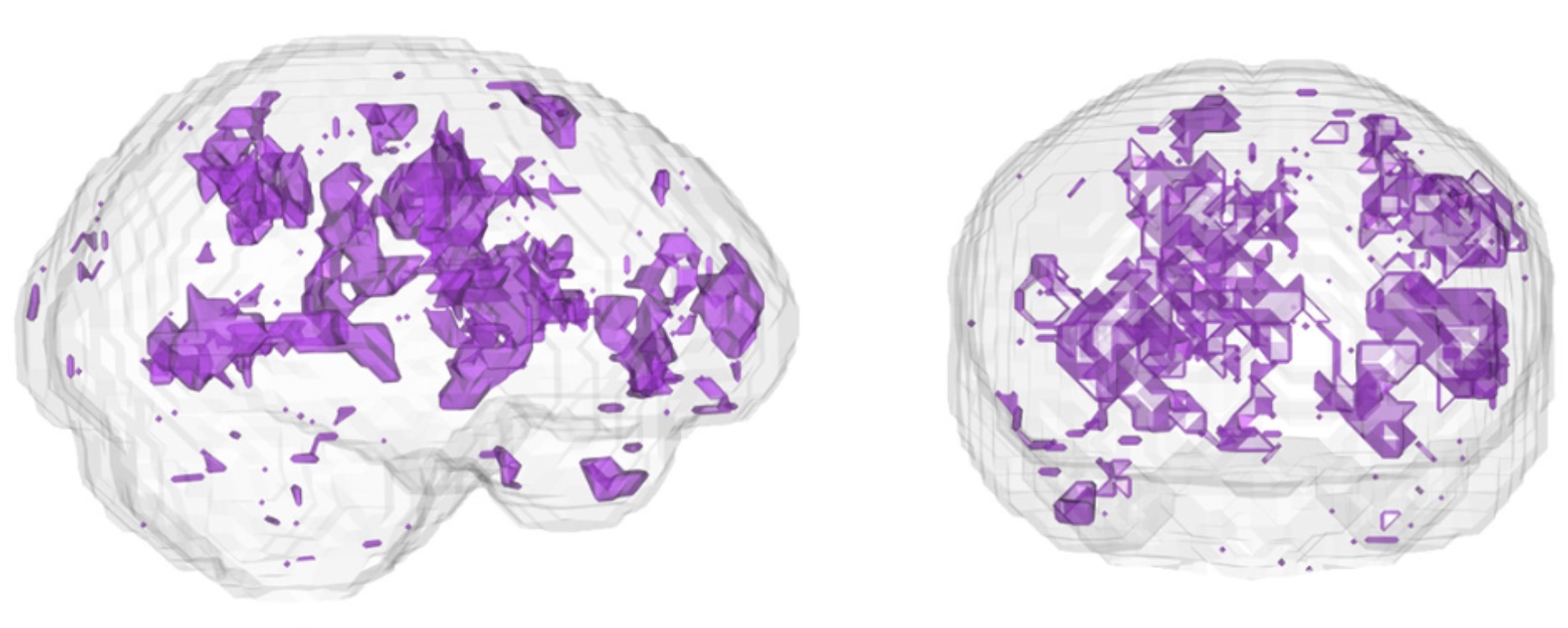
This data collection is the output of the modification of a regularised dimensionality reduction algorithm, Modified Locally Linear Embedding (MLLE). In comparison to other preprocessed collections, this output purports to contain the quantum fluctuations as measured by nuclear magnetic resonance, also known as functional Magnetic Resonance Imaging (fMRI). This methodology is motivated by the neurophysiological basis of the fMRI signal. There is voluminous evidence (fMRI studies) showing the haemodynamic response function (HRF) differs across different brain regions. Logothetis et al. [1] established important properties that link the fMRI signal to the underlying electrophysiological activity.
These reconstructions leverage the observations of Logothetis et al. [1] study by encoding a local structure at each voxel in the brain that is then globally optimised (MLLE). The various dimensionality parameters (d) reflect the ongoing investigation of the research. Our investigations have found that on and after a certain ‘d’ parameter (usually smaller than the original temporal dimensionality of the fMRI), the reconstructed fMRI significantly outperform the original and PCA-transformed fMRI, when using Leave One Out Cross Validation with sequential forward search to select volumes in the images in an incremental fashion.
[1] Logothetis, N., Pauls, J., Augath, M., Trinath, T., and Oeltermann, A.. Neurophysiological investigation of the basis of the fMRI signal. Nature 412, 6843 (2001), 150–157.
Download using Cyberduck
There are file transfer programs that can handle S3 natively and will allow you to navigate through the data using a file browser. Cyberduck is one such program that works with Windows and Mac OS X. Cyberduck also has a command line version that works with Windows, Mac OS X, and Linux. Step-by-step instructions for using Cyberduck are shown below.
The end result should appear as follows:
Download using Terminal
If you prefer to use AWS CLI, the s3 URI to this data is: s3://fcp-indi/data/Projects/ABIDE_Initiative/Outputs/lle/.
Information about naming convention and data structure can be seen HERE.
The following datasets were preprocessed using the C-PAC pipeline with the default settings:
- Yale
- UM_1
- UM_2
- Stanford
- USM
- UCLA_1
- Deobliquing, reorienting and skullstripping the T1 scan.
- Deobliquing and reorienting the functional volumes, followed by calculating the motion correction parameters using the mean over all functional volumes, and concluding by skullstripping these volume (slice_timing_correction=True).
- Non-linearly registering the anatomical volume to MNI T1 template space at 1x1x1 mm resolution. Note that this step uses the affine transformation matrix obtained from linear registration as a starting point.
- Registering the reoriented, deobliqued, motion-corrected and skullstripped functional volumes to MNI space using the registered anatomical obtained from the previous step, followed by resampling these volumes using the voxel size specified by functional scan parameters.
Note: spatial smoothing is disabled for all of our preprocessed data (i.e. inputs.linear_func_to_anat.no_resample=True inputs.linear_func_to_anat.no_resample_blur=True inputspec.interp = "sinc"). Detailed information about the preprocessed output from C-PAC (i.e., the complete output, including all intermediates with the generated metadata) is contained in tar files named by the site of collection: https://dx.doi.org/10.21227/zkkm-es92.
The following datasets were preprocessed using Statistical Parametric Mapping (SPM12) with the default settings (except where noted):
- Caltech
- CMU
- KKI
- Leuven_1
- Leuven_2
- MaxMun
- NYU
- OHSU
- Olin
- Pitt
- SBL
- SDSU
- Trinity
- UCLA_2
- Estimating the re-alignment parameters for the functional image (quality=1, fwhm=0, separation = 3), registering all functional volumes to the first volume (rtm=0, 3rd degree B-spline interpolation, no wrapping or weight).
- Estimating the coregistration parameters by registering the first volume of every subject's functional image as the source, the subject's anatomical volume as the reference, and the remaining functional volumes as the other images.Settings were otherwise default, except separation which we set to [4 2 1].
- Estimating the warping parameters for affine registration of the subject's anatomical volume to MNI T1 template space at 1x1x1 mm, with "no correction" for the Bias FWHM, a sampling distance of 2, and smoothness set to 0.
- Resampling the subject’s functional volumes according to the warping parameters determined in step 3, without a bounding box, the voxel size set to the same as the raw functional volume, and 4th degree B-Spline Interpolation.
- Consistent with the ABIDE initiatives, none of the datasets were checked for movement.
- Earlier datasets (UM_1, UM_2, UCLA_1, USM, Stanford) preprocessed with C-PAC underwent very rudimentary checks (at the time of preprocessing).
- List and notes about subjects not included:
- ABIDE 1:
- OHSU: subs 0050155 and 0050165 do not have rest_1, so rest 3 was used in those cases.
- UCLA 1: 0051232, 0051233, 0051242, 0051243, 0051244, 0051245, 0051246, 0051247, 0051270, 0051271 were not reconstructed since their anatomicals is not included.
- UCLA 2: 0051310 is missing anatomical, so it could not be reconstructed.
- UM_2: 0050383, 0050403, 0050413, 0050422, 0050427 were not reconstructed because the preprocessed image is warped.
- ABIDE 2: The following anatomical or functional volumes are not included:
- ETH_1: 29069, 29081
- IP_1: 29622
- UCD_1: 30004
- ADHD200: The following anatomical or functional volumes are not included:
- NYU: 0010016, 0010027, 0010055, 0010098, 0010105, 0010127
- NeuroIMAGE: 3808273
- WashU: 0015011, 0015018 (do not have a 'paired' 'run-1' for functional/anatomical for any of the sessions)
For questions please contact gagan[at]g-a[dot]ca.
If you use data from this LLE Preprocessed repository, a citation would be appreciated but it is not compulsory: